Extended usage¶
Create a new Pecube project¶
To create and run a new Pecube project, go to New input file or press Crtl+N (3, see Introduction), a window pops up and asks you to provide the name of the new project (Figure 1). After clicking the “Ok” button (Figure 2), you will be able to provide and set all of the Pecube input parameters for your project.
Figure 1. Enter a new project name. This window shows up when clicking on the “New input file” action.¶
The window should look like Figure 2. On the top left, you could find and access all the projects you have loaded into the interface, and thus navigate through their respective parameters. The right part of the interface will host extra input parameters described in the next sections.
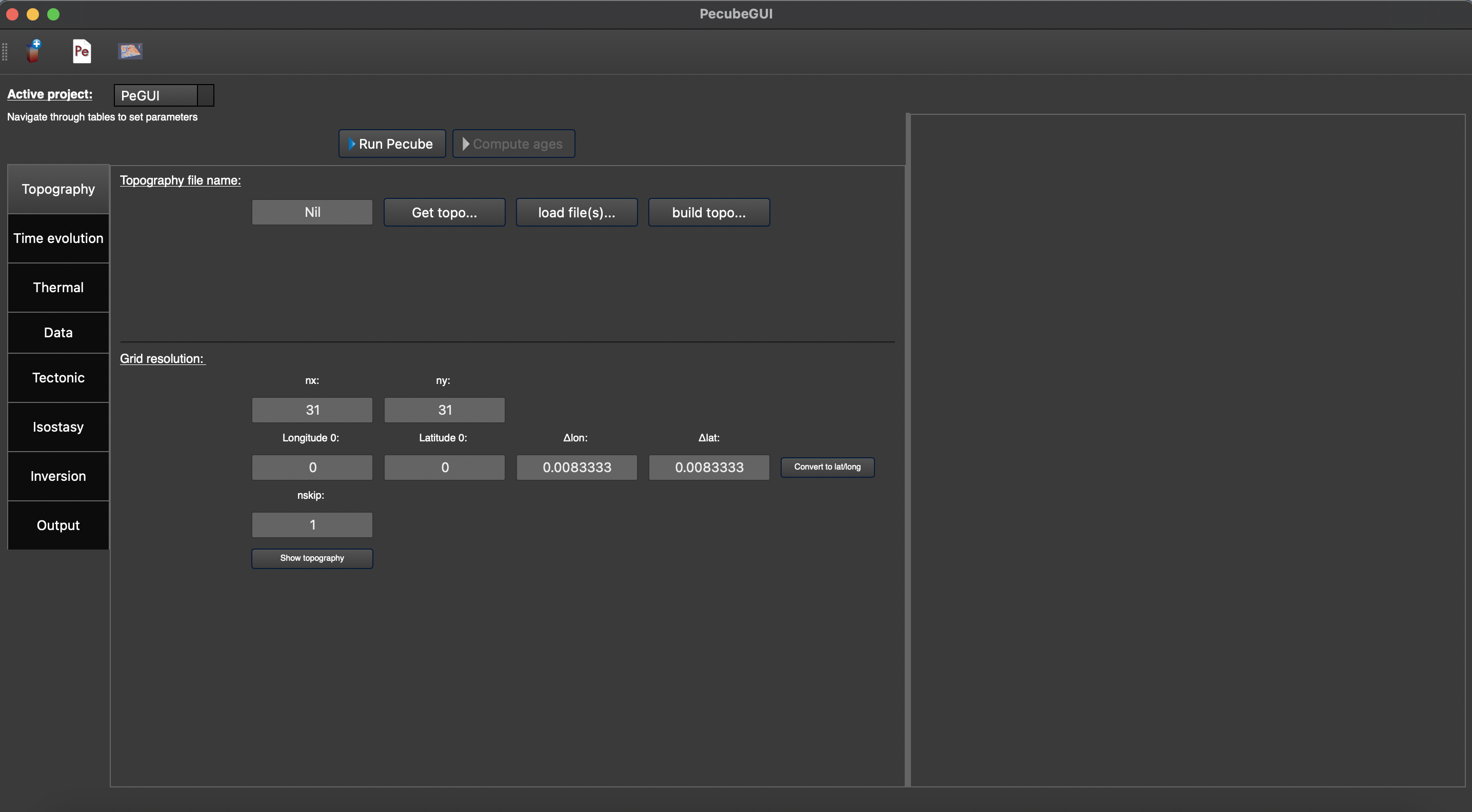
Figure 2. Tab to set up the input parameters for Pecube. On the top left, you can check the name of your project (here: ‘PeGUI’)¶
The input parameters are organised into categories similar to what is referenced in the Pecube’s user guide. All the parameters are sorted in 8 categories: Topography, Time evolution, Thermal, Data, Tectonic, Isostasy, Inversion, and Output parameters. For more details on the meaning of each input parameter please refer to the Pecube’s user guide. However, you could access to a short description of each parameter by simply flying the mouse cursor over the parameter labels. After one second a text should appear and describe the parameter.
In the following, I provide a description of all the tabs, and how to provide the input parameters in PecubeGUI.
Provide input parameters¶
Topography tab¶
a csv file containing one column of elevation
a serie of output files from a spm to be read by Pecube (see Pecube user guide – “Topography parameters”)
a DEM file (i.e., raster file ‘.tif’)
a DEM extracted from an interactive map (“Get topo…”, Fig. 2)
You can also choose to build a simple sinusoïdal topography (‘Build topo…”, Fig. 2).
For the three first points, click on ‘load file(s)’ (Figure 2) will allow you to select the file(s) to load. A window pops up. The figure 3 shows an example where a serie of files has been loaded from a spm like iSOSIA [Egholm_et-al-2011].
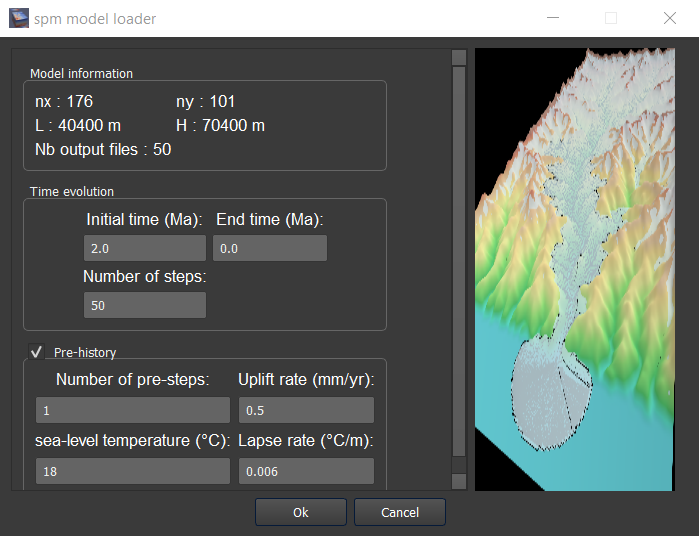
Figure 3. Spm loader window.¶
number of pre-steps: how many steps for the pre-spm history you want to provide topographic information
uplift rate: this will be assumed to be spatially uniform and applied to all pre-history steps
sea-level temperature
atmospheric lapse rate
Important
If you use the DEM from the SRTM data hosted by OpenTopography, please use this citation: NASA Shuttle Radar Topography Mission (SRTM) (2013). Shuttle Radar Topography Mission (SRTM) Global. Distributed by OpenTopography. https://doi.org/10.5069/G9445JDF. Accessed: 2022-11-18. With the in-text citation: NASA Shuttle Radar Topography Mission (2013).
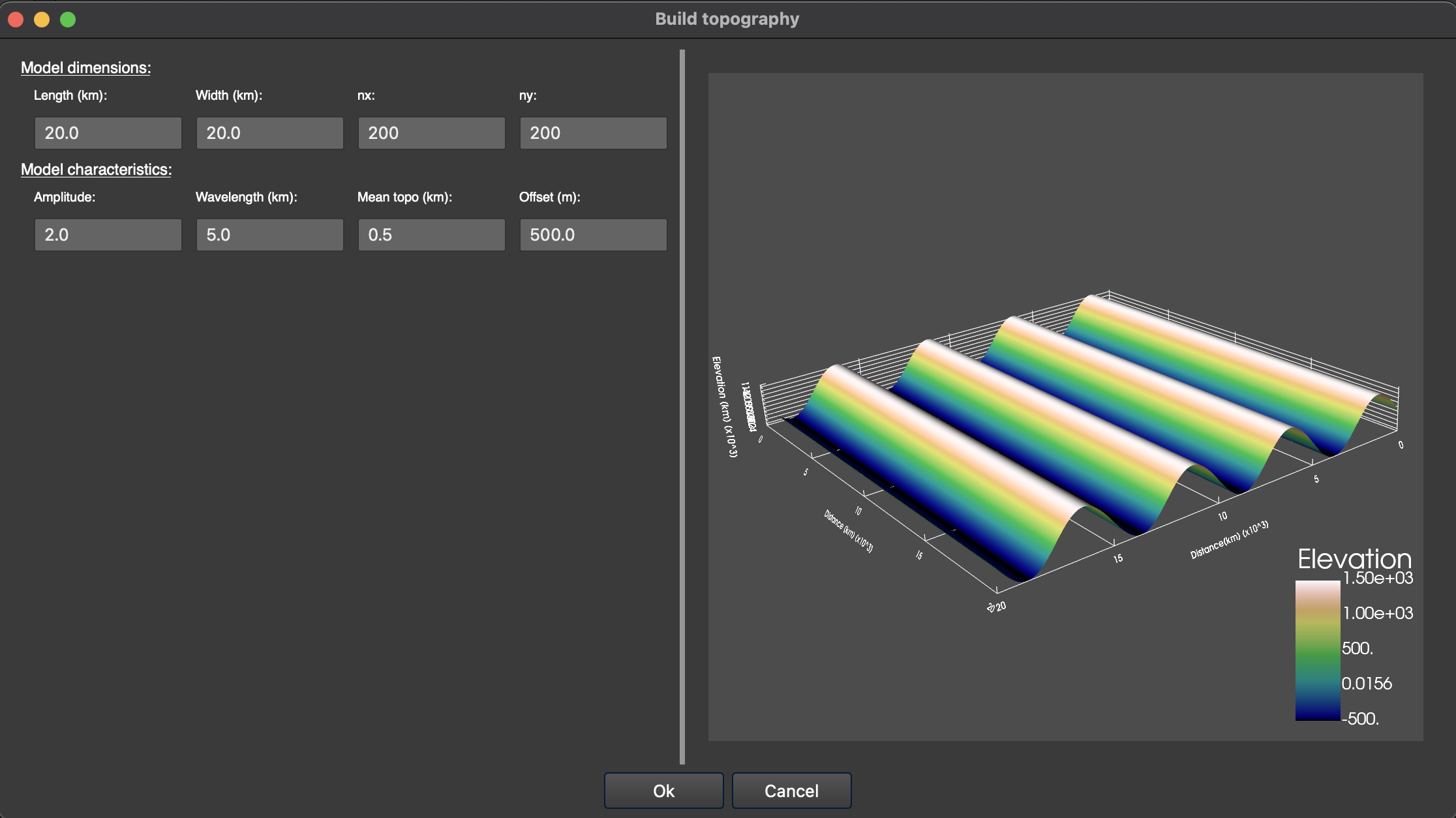
Figure 5. Window to build a synthetic sinusoïdal topography.¶
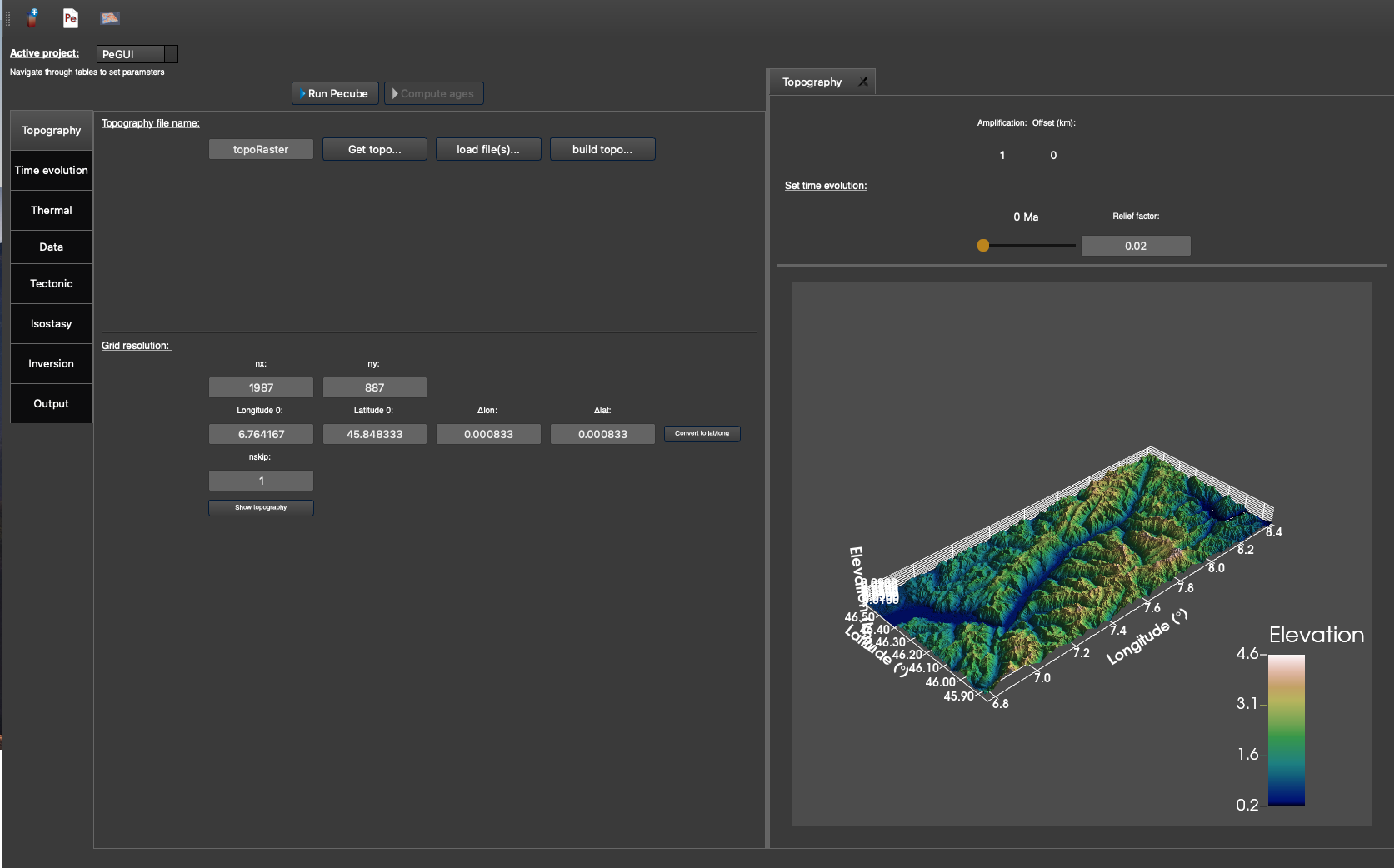
Figure 6. “Topography” tab with the topography shown on the right-hand side, after clicking the “Show topogaphy” button. The topography is loaded from a DEM from the Rhone valley in Switzerland.¶
Time evolution tab¶
In this tab (Figure 7) you can provide all the parameters that control the time evolution of the input topography. In PecubeGUI you can provide the time evolution parameters (cf. “time_topo”,” amplification”, “offset”, and “output”) by filling in the table or by copying/pasting values from an excel file to the table. The number of rows in the table automatically updates from the value written in the parameter “ntime” (Figure 7).
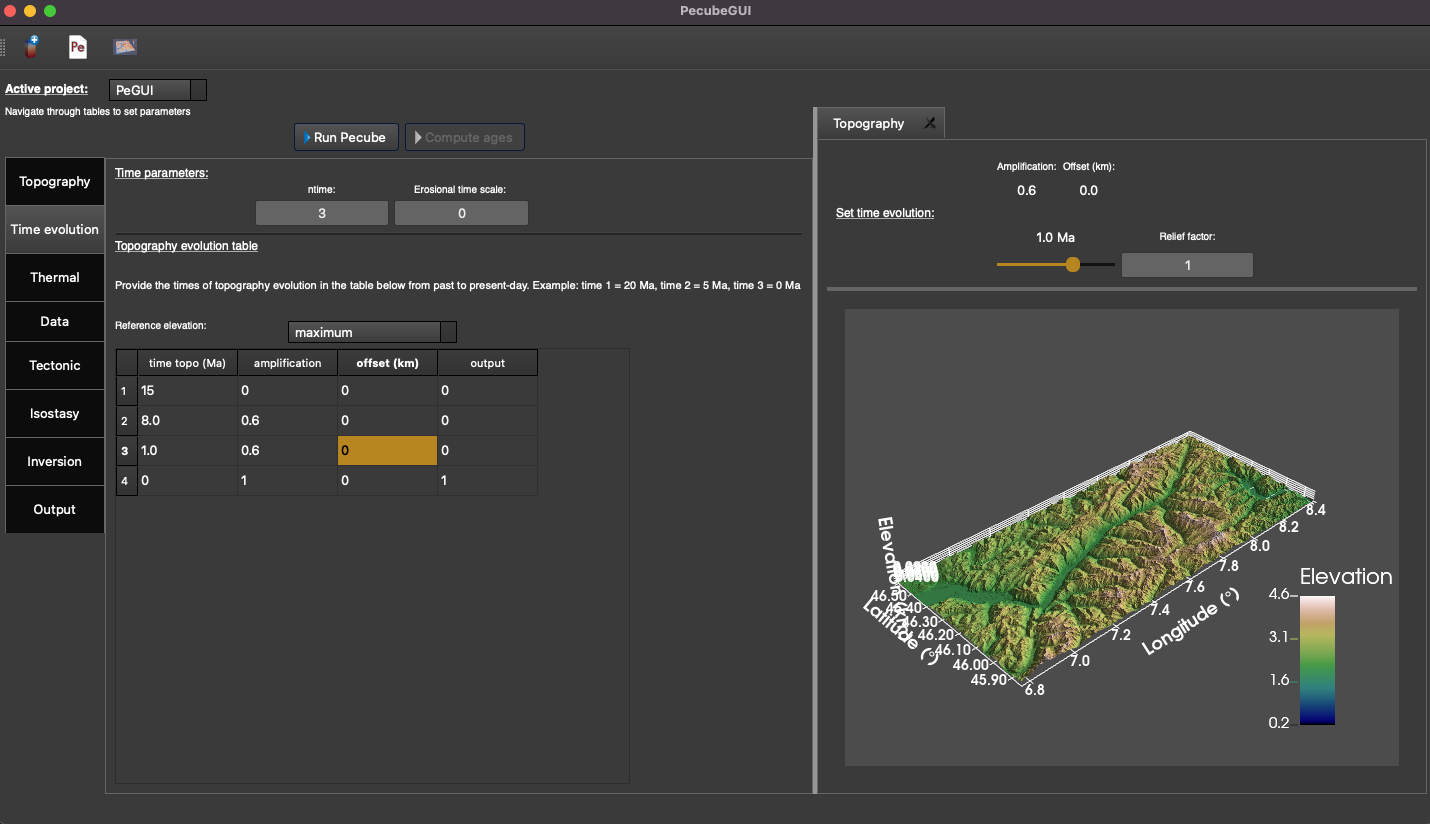
Figure 7. “Time evolution” tab where to provide the parameters related to the time evolution of the topography. In this example, the topography evolution is defined relative to the maximum elevation.¶
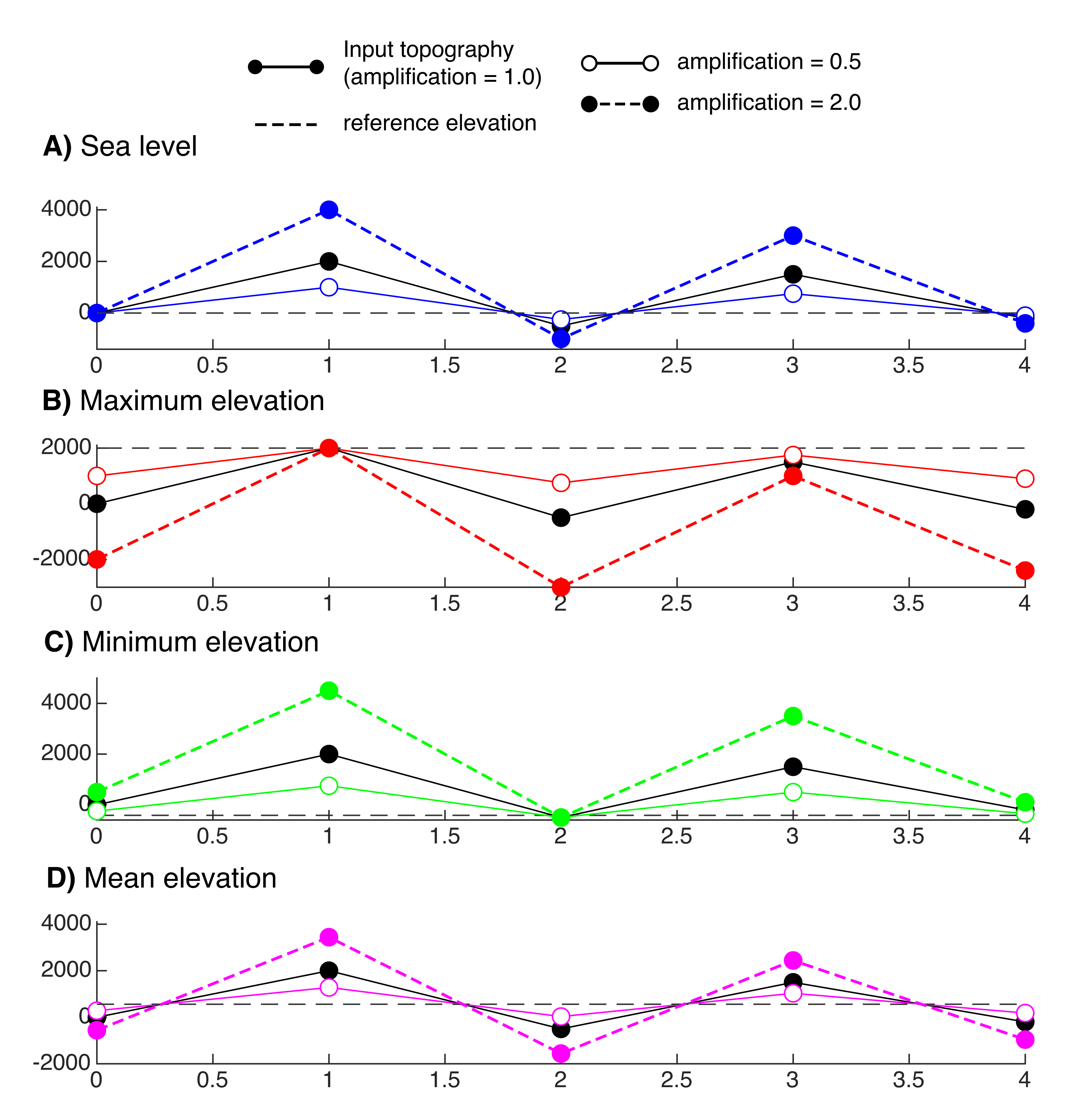
Figure 8. Reference elevations from which to compute the topographic evolution. These references are A) sea level, B) minimum, C) maximum, D) mean elevation.¶
Thermal tab¶
The user can also set a non-uniform heat production rate. An exponential decrease of heat production rate with depth can be specified by checking the box “use e-folding HP”. A small window pops up where you can provide the heat production rate and the e-folding depth. In the input parameters for pecube, the heat production rate will then be negative to inform pecube you want to use a non-uniform heat production rate.
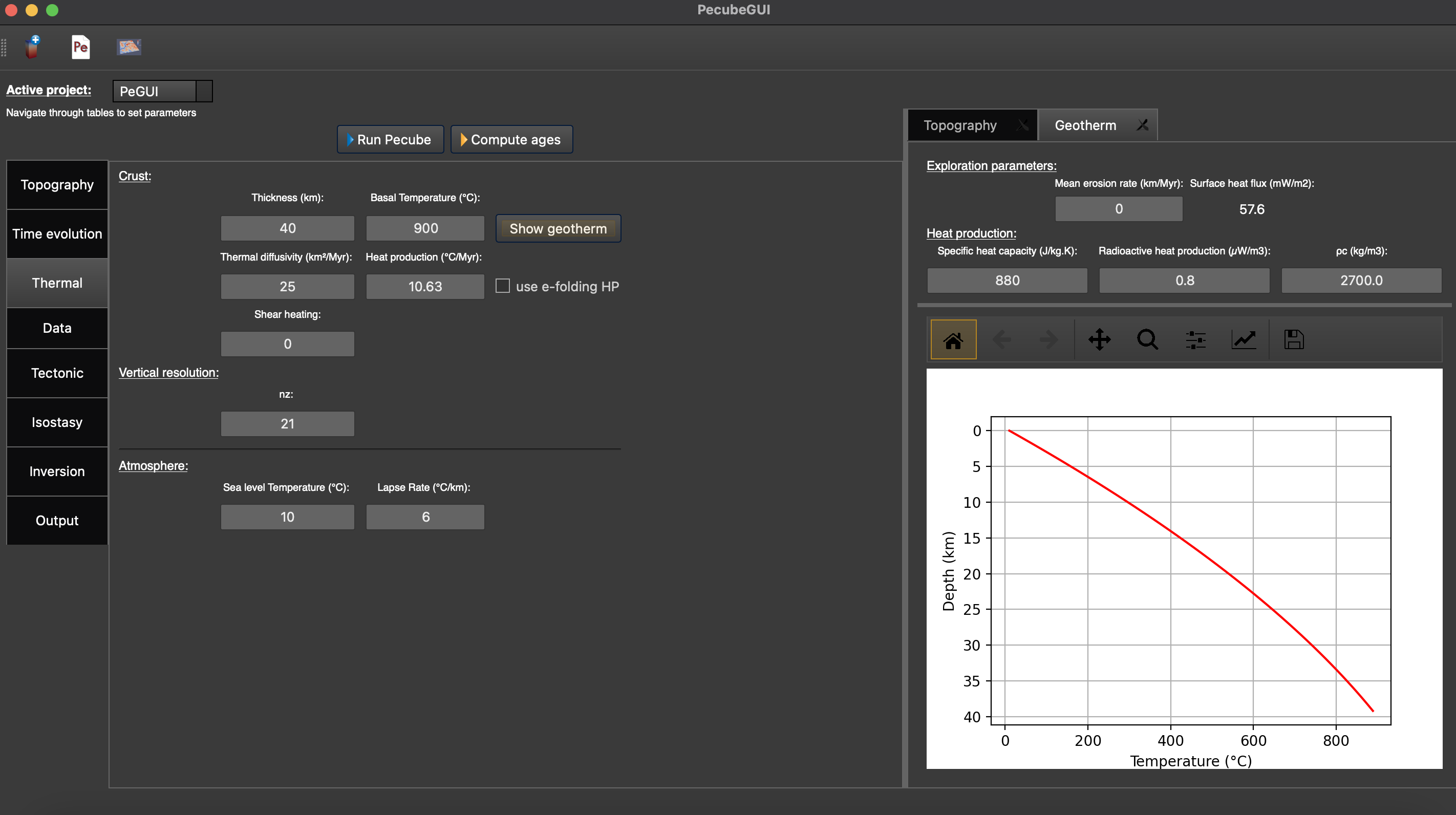
Figure 9. “Geotherm” tab where to provide parameters related to Thermal properties of the crust and the atmosphere.¶
Data tab¶
none: Pecube will not predict any thermochronological ages
for all nodes: Pecube will predict thermochronological ages for all nodes at the surface of the Pecube model.
sample specific: Pecube will predict thermochronological ages only for specific sample locations provided by the user.
Warning: the ID of a sample must not include space ! Finally you can check for your sample location on the input topography by clicking on “Check sample locations”, as well as the predicted elevation of the samples on the input DEM (potentially degraded wit ‘nskip’ parameters) by clicking on ‘Check sample elevations’.
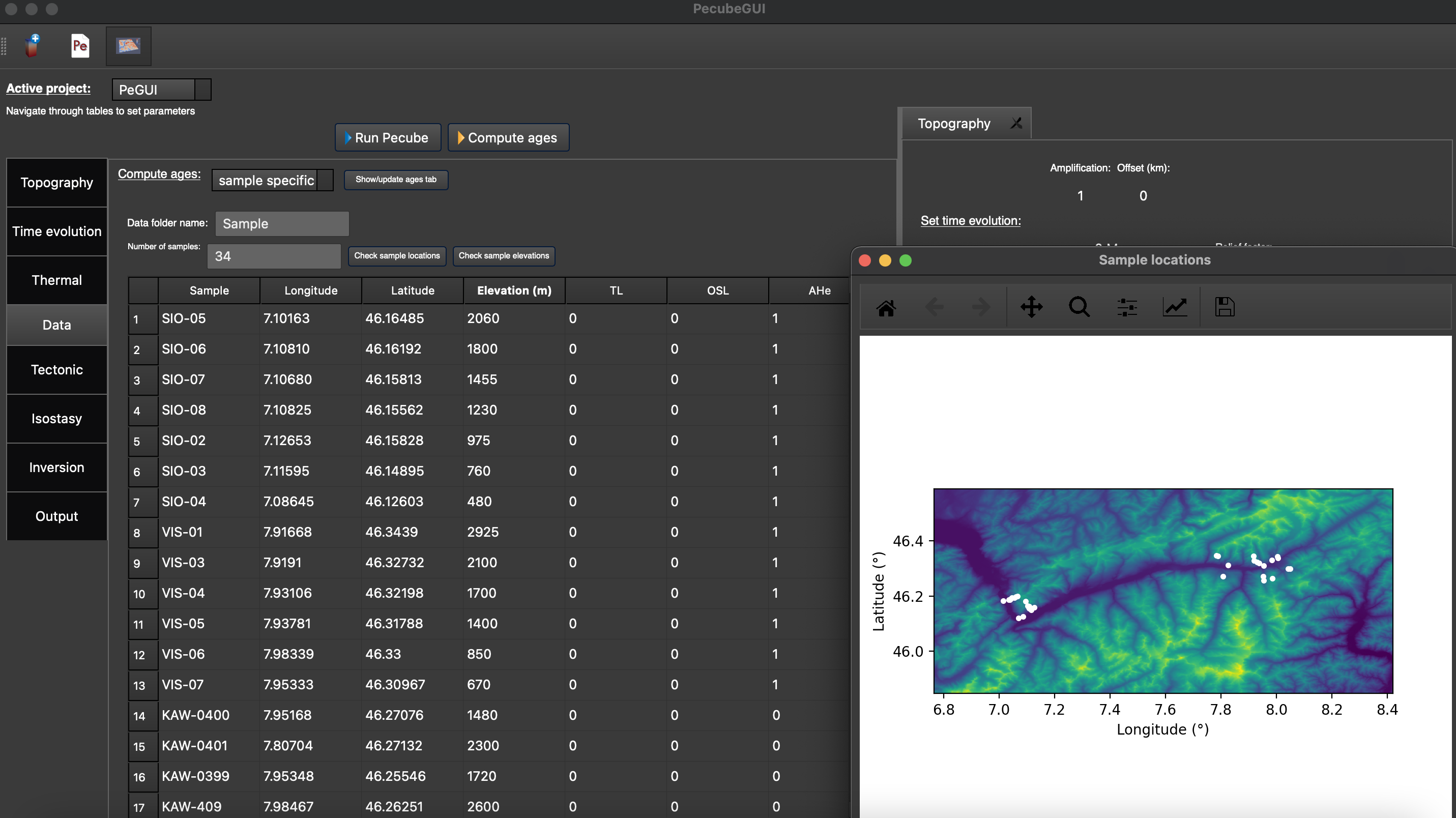
Figure 10. “Data” tab where to provide the sample location(s) and number of observations for each thermochronometers. The extra window shows the location of the samples, here in the Rhone valley area (data from Valla et al., 2012)¶
After providing the number of observations, you can click on ‘Show/update ages tab. A tab will open, where you can provide information for each thermochronometer (Figure 11).
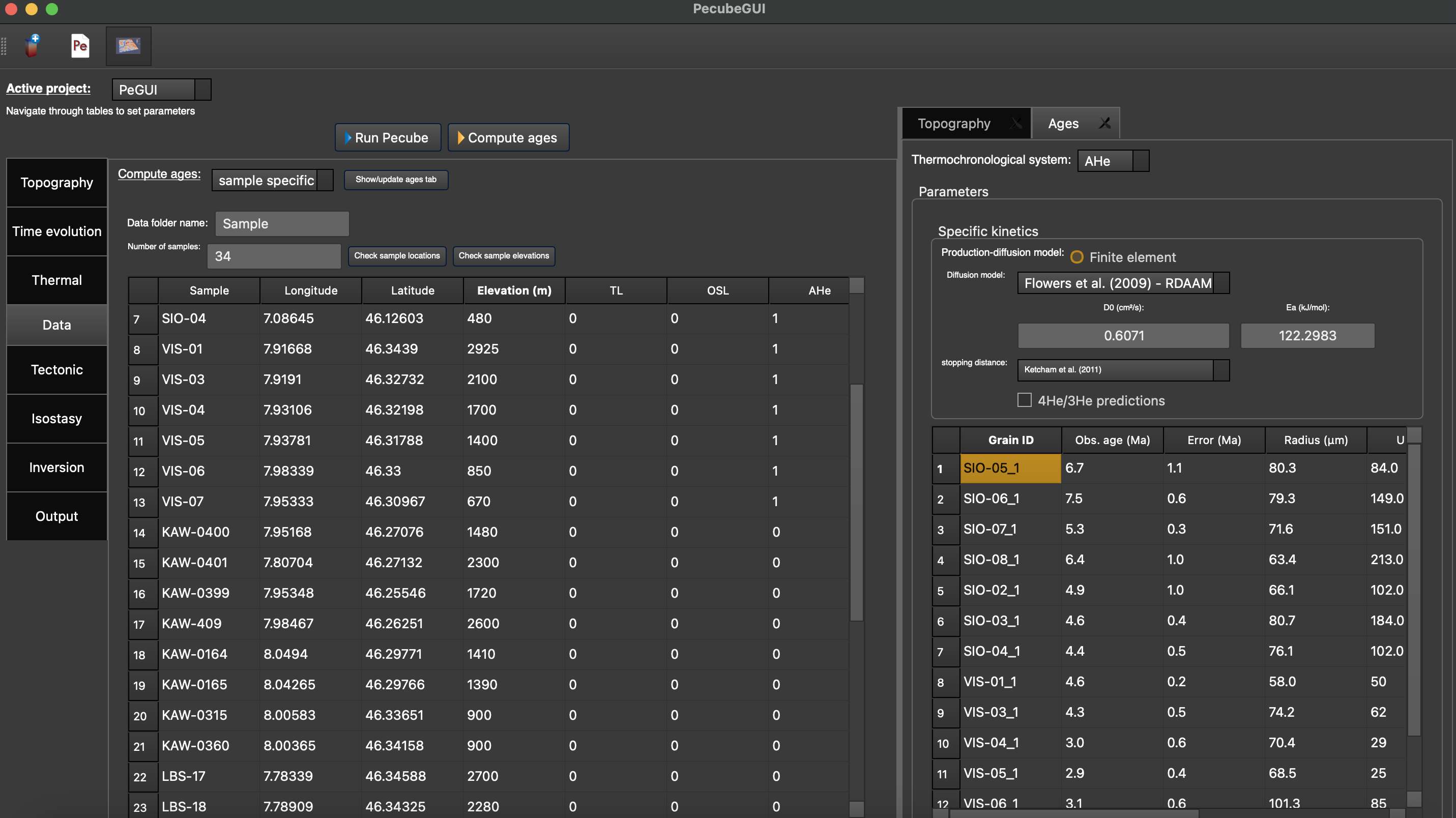
Figure 11. “Ages” tab where to define the thermochronometers to use. Here, the example is made with sample specific predictions for the apatite (U-Th)/He system.¶
Diffusion model: the helium diffusion model to use. The options are the Farley et al. (2000), Shuster et al. (2006), and the radiation damage models from Gautheron et al. (2009), Flowers et al. (2009, RDAAM) and Willett et al. (2017, ADAM).
Ea: The activation energy (kJ.mol-1). This is automatically updated according to the selected diffusion model, but it can be changed at the user’s discretion.
D0: the diffusivity parameter value for infinite temperature (cm2.s-1). The value updates according to the selected diffusion model.
stopping distances: stopping distances for alpha particules from Farley et al. (1999) or Ketcham et al. (2011).
Table of observations: The table includes the observed ages and their uncertainties, the size (radius) of the grains, their uranium and thorium concentration (in ppm), and the rmr0 kinetic parameters (only for Flowers et al. (2009) and Gautheron et al. (2009) diffusion models). In the current version, the grain is assumed spherical.
4He/3He predictions: allows to predict 4He/3He profiles for each grain. When checked, a new window opens. Within this window, you can provide your heating schedule, with the number of steps, or let the default heating schedule. This will be used in the diffusion model to simulate a degassing experiment and compute 4He/3He ratios. The heat is in °C and the duration in hours. The same heating schedule is used for each grain.
Tectonic tab¶
no uplift: compute the effect of topographic change only on thermal field
bloc uplift: vertically uniform movement of the entire domain
faulting: slip rate along fault(s)
Note
To define the type of fault, we keep on the convention described in the Pecube’s documentation. To define a thrust the velocity has to be negative, a positive velocity means a normal fault (i.e., ‘velo’ in the table).
The order you define the position of the points of the fault(s) matters. The convention is that the fault geometry is defined to the right of the strike of the fault (see Pecube’s documentation for more details)
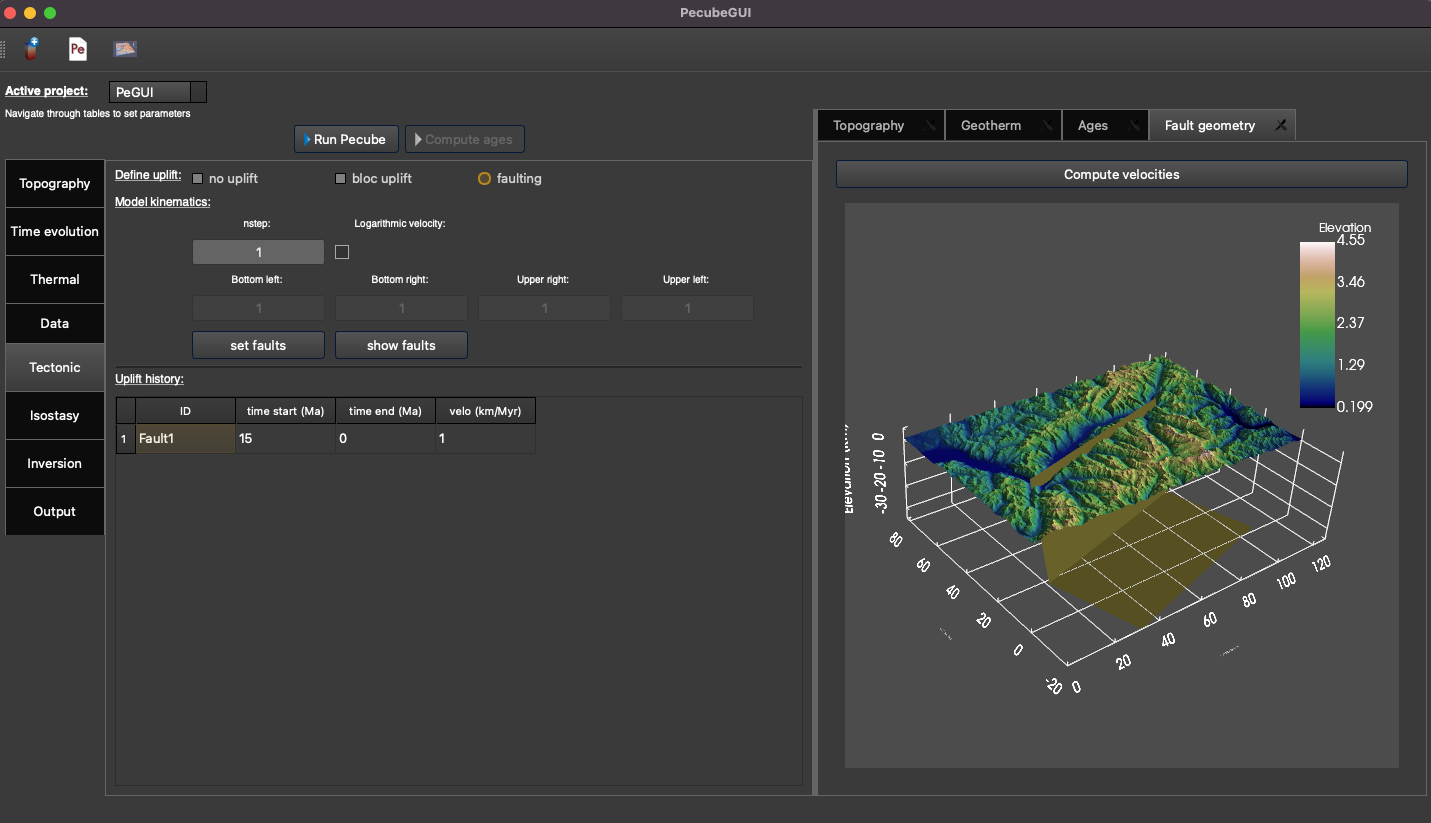
Figure 12. “Tectonic” tab where to provide parameters related to kinematic of rock uplift.¶
Run a Pecube model¶
Note
When several projects are opened, the consoles are gathered in a single window to have a quick overview of all the running simulations.
Plotting results¶
In that section, I provide an overview of the chart part of PecubeGUI. There, you can plot results from your Pecube run.
Plot 2D data¶
To plot 2D data in PecubeGUI, first switch to the chart’s window by clicking on ‘show ouput’ (see Figure 1 in “Introduction”, n°5). You should see the window shown in Figure 13. On the left-hand side, you will find two tabs: Data and Properties. The first tab enables to load new data:
Load project…: load a Pecube input file to plot data from that project.
Add 3D model…: load a vtk file to render a 3D model.
Remove data…: remove one or several plots. To do so, on the plot list on the left-hand side of the interface, select the plot you wish to remove and click ‘Remove data…’.
Further data…: a list of 2D data you can plot.
In the current version, and depending on your input parameters, Pecube can output several files. These files are:
CompareAge.csv: This file contains the predicted and observed ages as well as sample ID and coordinates.
TimeTemperature.csv: stores the thermal path of each sample location you provided, either by writing directly in a file in the ‘Data’ directory of your project, or by specifying the locations in PecubeGUI when want to predict specific AHe ages (see ‘Output parameters’ tab). For this file to be created, you also need to check ‘save PTT paths’ in the ‘Output parameters’ tab.
CoolingRates.csv: contains the time-temperature paths from all nodes in the model. This file is created if the option “Cooling rates” is checked (see Ages-tab). This allow the user to plot a 2D map of cooling rates defining a temperature or time interval.
PecubeXXX.vtk: This file is located in the “VTK” directory of your project. If loaded for 2D data plot, a window will show up and ask you which data to plot from the file. You can extract, for instance, the 2D spatial distribution of the temperature at a specified depth, or extract the depth of an isotherm.
AgeXXX.vtk: This file is located in the “VTK” directory of your project. If loaded for 2D data plot, you can choose to plot the 2D spatial distribution of the erosion rate or the predicted ages, at the surface of the Pecube model (only with the “for all nodes” option, see Ages-tab).
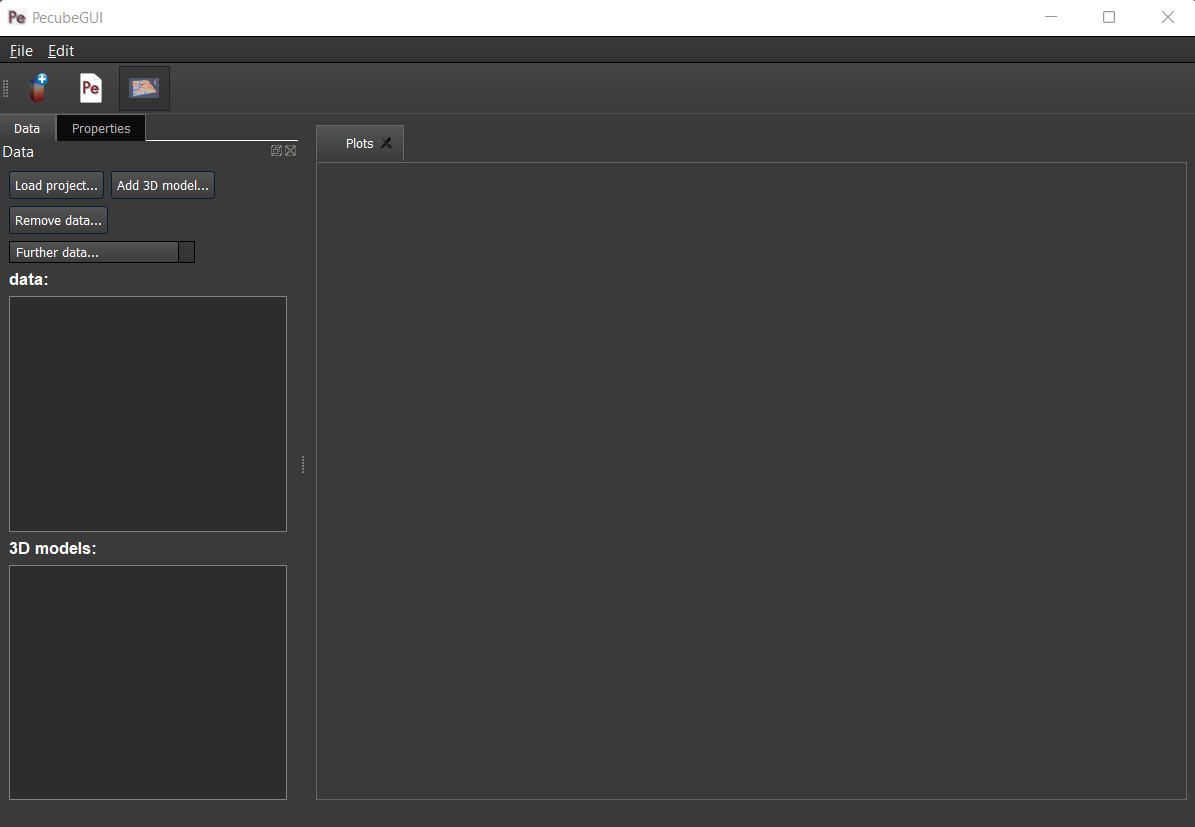
Figure 13. Chart’s window.¶
For each Pecube project, the 2D data you can plot will be listed in “Further data…” (Figure 13). However, you first need to tell PecubeGUI which pecube project you want to work with. To do so, click on ‘Load project…’, then a window appears and ask you to choose an input file corresponding to the Pecube project you desire to plot from. After loading the input file, the list below will update and show you what kind of data you can plot. From there you could plot:
Age-elevation: the ages plotted against elevation. If you computed ages for all surface nodes of the model, then you will be asked to choose at which time step(s) you want to plot data. If you computed ages at specific locations and for several thermochronometers, all of them will be plotted along with observed data you provided. Then you will be free to show/hide data as you wish (see Figure 14).
Date-eU: plot ages against effective uranium. Works only if you computed AHe ages at specific locations.
Age-comparison: plot observed vs predicted ages.
Age transect: plot observed and predicted ages along transect (Latitude, longitude, or projected).
Tt paths: plot the thermal path of each samples. Works only if you computed ages at specific locations.
4He/3He data: plot either 4He/3He spectra or step ages profiles.
2D map of cooling rates: compute cooling rates for all surface node of the model. You will be asked to define the temperature or time range on which you wish to calculate the cooling rates, as well as the interpolation method you want to use.
2D map of temperatures: plot the temperature/depth map at a certain depth/isotherm. To plot this map you will need to load one of the “PecubeXXX.vtk” file in the “VTK” directory of you pecube project.
2D map of Ages: plot the ages at the surface of the model. Works only if you computed ages for all surface nodes! To plot this map you will need to load one of the “AgesXXX.vtk” file in the “VTK” directory of you pecube project.
Inversion results: plot the result of inversion. Three plot options are possible: 1) 2D parameter space, plot parameter X vs parameter Y in scatter plot where colors represent the misfit value. This option is enabled when running the sampling stage of the Neighborhood Algorithm (included in Pecube). The two other options 2) 2D parameter space + 1D PDF, and 3) 1 PDF single parameter, can only be used if the second stage (i.e., the appraisal stage) of the Neighborhood Algorithm has been run and the file “nab.out” is in the “NA” directory of your Pecube project.
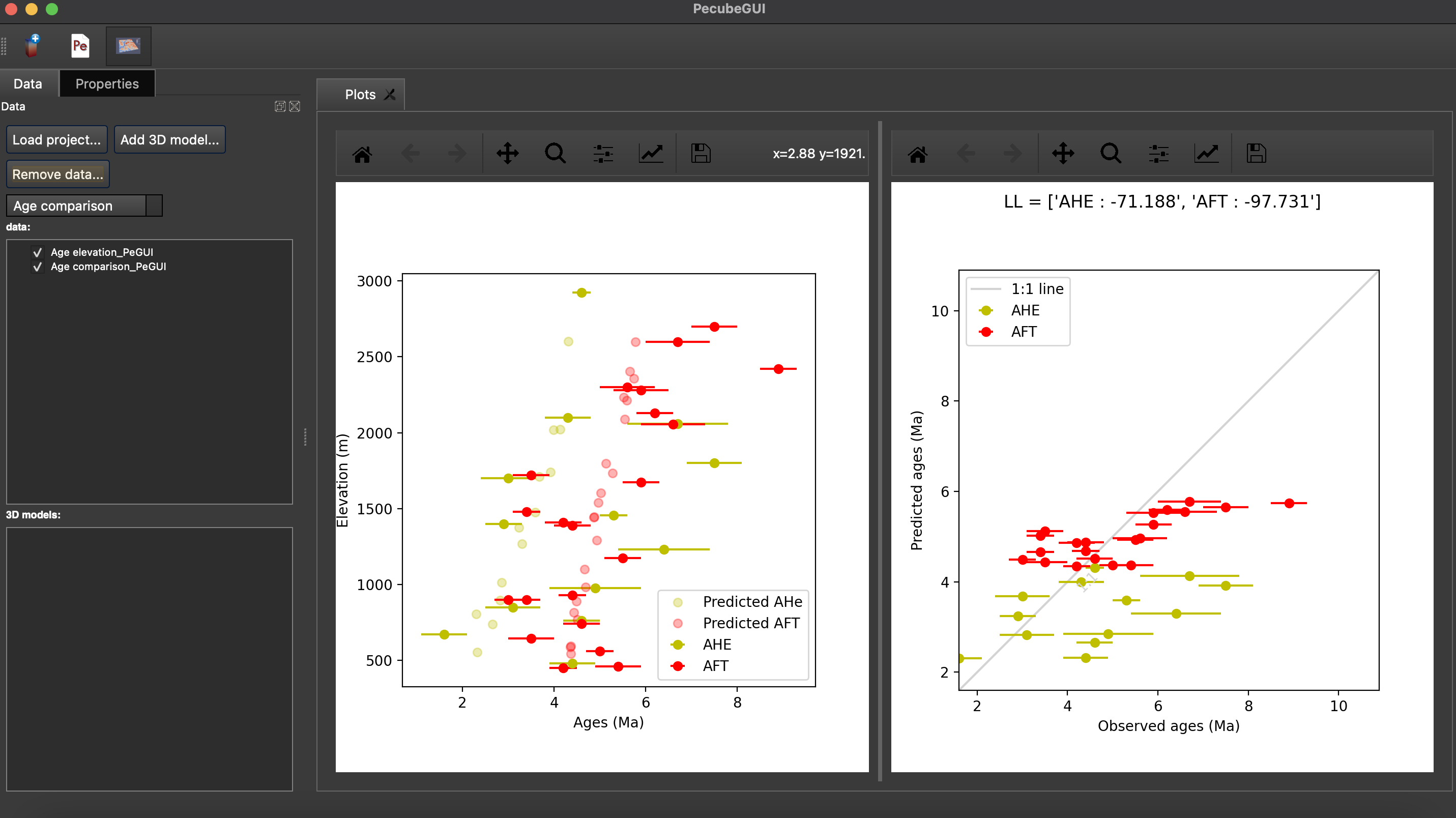
Figure 14. An example of an age-elevation plot. Here, AHe and AFT ages are predicted and compared against observations from the Rhone valley area.¶
Note
When plotting predictions from specific locations, and if observed data are provided, a misfit criteria between predicted and observed data is shown on the plot. This misfit criteria is known as the likelihood (in fact log-likelihood) that is the probability to have the observed data according to the model predictions. The log-likelyhood is defined following Braun et al. (2012): \(LL = -\sum_{j=1}^{N}(\frac{ln(2\pi)}{2}+ln(\sigma_j)+0.5(\frac{S^{obs}_j - S^{pred}_j}{ \sigma_j})^2\).
Where \(S^{obs}_j\) the observed data j and \(S^{pred}_j\) the predicted data j, \(\sigma_j\) the error on the observed data j, and N the total number of observed data. The higher the value of LL, the better is the match between observed and predicted data.
Visualize 3D data¶
Data range: set the range of data for the colorbar.
Current data: list to choose the data to show (i.e., for the colormap).
X, Y, Z scales: to scale the 3D model in the x, y, and z, directions.
Reset camera position: reset the camera view to the initial position.
Clear plot: remove the 3D model from the 3D interface.
Export image…: save a screenshot of the 3D interface.
Show box: to show the axes of the 3D model.
Show sample location(s): to show/hide sample locations within the 3D interface.
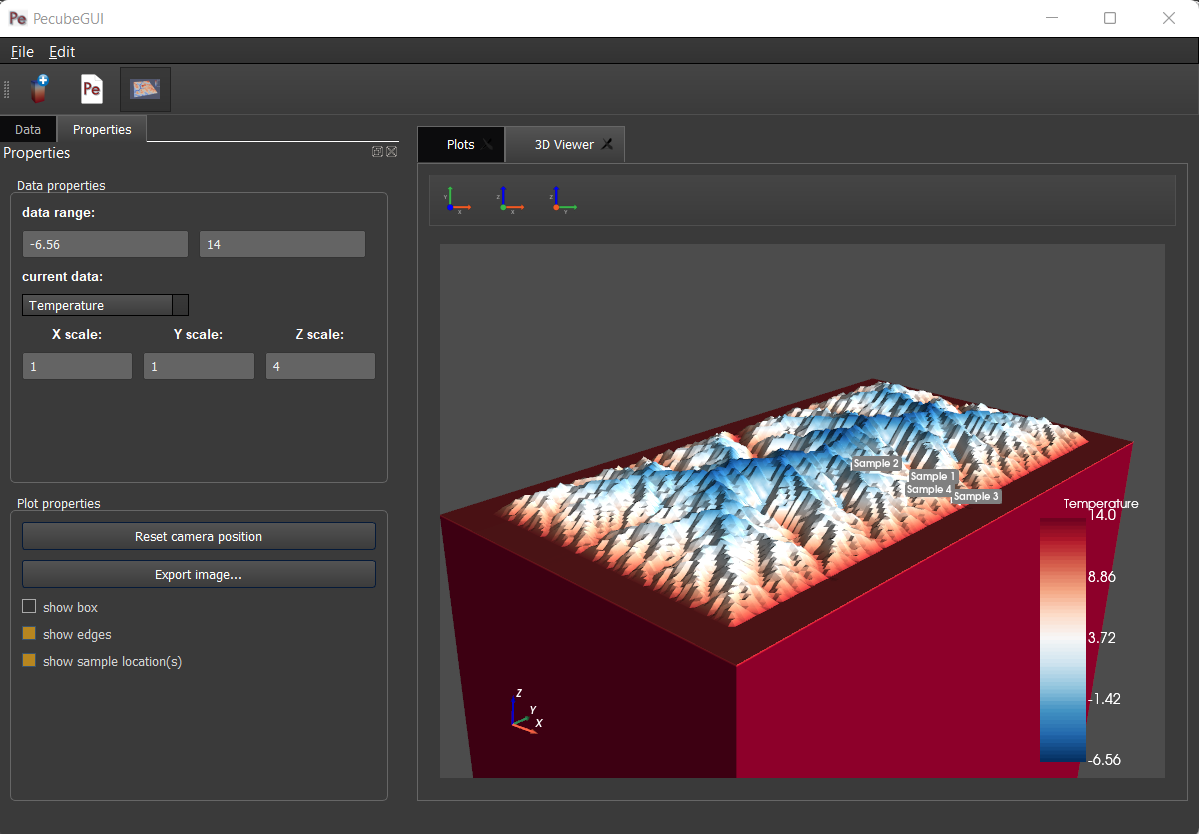
Figure 15. 3D viewer in PecubeGUI. An example is shown where the surface temperature is shown on the topography alongside with the sample locations that have been defined (see output tab).¶
References¶
Egholm, D. L., Knudsen, M. F., Clark, C. D., & Lesemann, J. E. (2011). Modeling the flow of glaciers in steep terrains: The integrated second‐order shallow ice approximation (iSOSIA). Journal of Geophysical Research: Earth Surface, 116(F2).
Reiners, P. W., & Brandon, M. T. (2006). Using thermochronology to understand orogenic erosion. Annu. Rev. Earth Planet. Sci., 34, 419-466.
Gautheron, C., & Tassan-Got, L. (2010). A Monte Carlo approach to diffusion applied to noble gas/helium thermochronology. Chemical Geology, 273(3-4), 212-224.
Ketcham, R. A. (2005). Forward and inverse modeling of low-temperature thermochronometry data. Reviews in mineralogy and geochemistry, 58(1), 275-314.
Sullivan et al., (2019). PyVista: 3D plotting and mesh analysis through a streamlined interface for the Visualization Toolkit (VTK). Journal of Open Source Software, 4(37), 1450, https://doi.org/10.21105/joss.01450
Piper, M. (2021). CSDMS Topography data component (Version 0.3.1) [Computer software]. https://doi.org/10.5281/zenodo.4608653